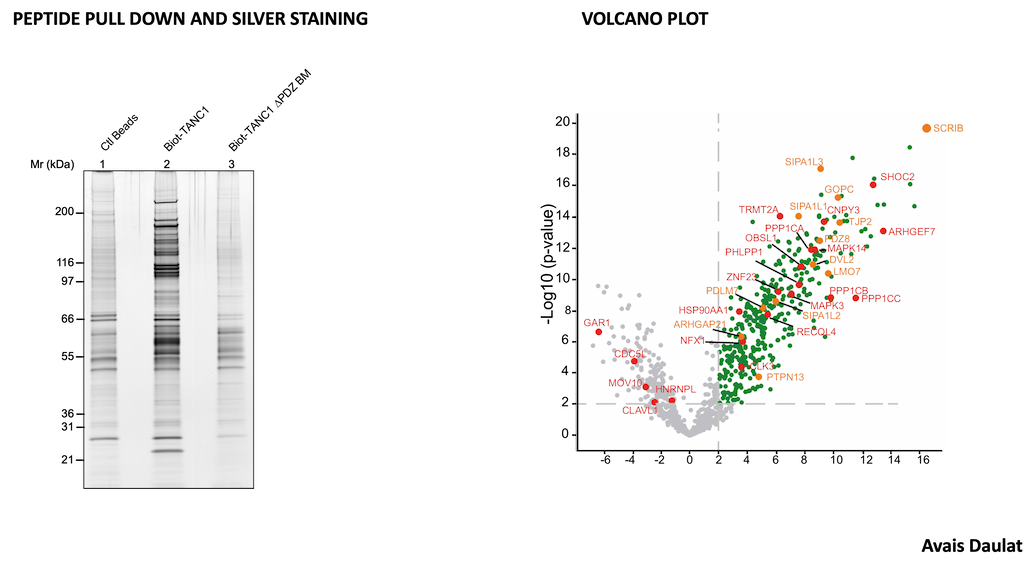
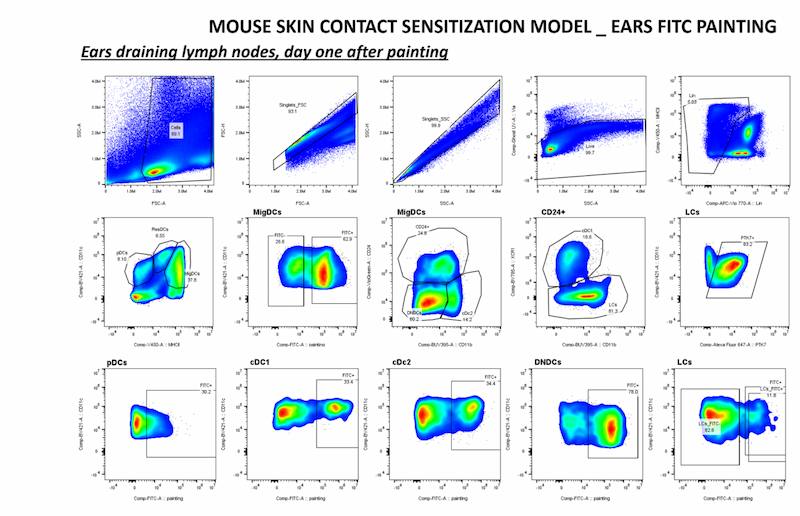
Our Targeting Signalling Networks & Micro-environnement in Cancer team studies cell signaling in cancer to identify vulnerabilities that will be the targets of tomorrow's drugs.
Studies are carried out using complementary strategies combining molecular, biochemical and multi-omics as well as cellular approaches, complemented by the use of ex vivo and in vivo models recapitulating the complexity of the tumor process. Our studies also benefit from interdisciplinary methodologies including computational biology, mathematical modeling and physics.
We explore signaling pathways in poor-prognosis cancers, from tumor initiation and throughout progression, exploiting vulnerabilities for the development of targeted therapies.
The team's themes revolve around two main areas:
Axis 1: Membrane signaling pathways in tumor cells.
We focus on poorly characterized signaling pathways in cancer at the molecular and functional levels. Importantly, these signals represent potential new targets against which we are developing bioactive molecules.
We explore the following aspects in particular:
- the molecular organization of signaling pathways,
- the molecular basis of cancer initiation and progression (cell dissemination, collective cell migration, treatment resistance, etc.) and the tumor microenvironment;
- the generation of inhibitor molecules with therapeutic potential.
Area 2: Modeling and targeting interactions between cancer cells and their tumor microenvironment: towards translational perspectives.
This axis explores the involvement of novel genes/mechanisms involved in the dialogue between cancer cells and their microenvironment, in particular immune cells. Our strategy is based on the combination of data from patients and ex vivo cancer models (organoids/tumoroids) derived from tumor samples, studied at the single cell level, and in vivo.
The projects
We are exploring signaling pathways whose functions and mechanisms of action remain largely unknown. These pathways could represent potential targets for new anti-cancer treatments. Our areas of research include:
- Mechanistic relationships between multiple signaling networks.
- The regulation of biological processes in cancer cells (initiation, dissemination, collective migration, resistance to treatment, etc.) and their microenvironment.
- The design of optimized strategies for generating blocking agents.
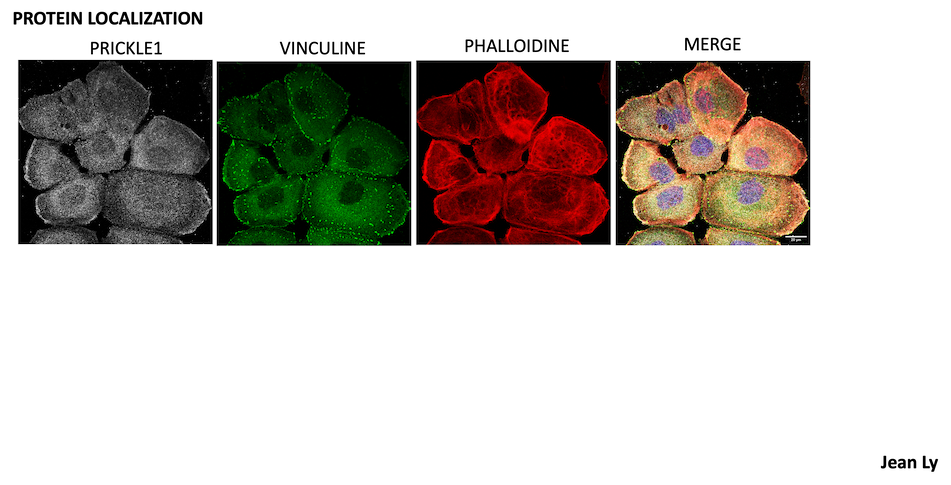
The WNT/Planar Cell Polarity (PCP ) signaling pathway plays a key role in the early and late stages of cancer. We have documented its mode of action in aggressive cancers and explored strategies to target its membrane or sub-membrane components.
1.1 VANGL2 is a WNT/PCP pathway receptor associated with poor prognosis in triple-negative breast cancer (TNBC). Our recent work has shown :
- Co-purification of VANGL2 with a GPCR receptor of the WNT/PCP pathway.
- An association between expression of this GPCR and reduced metastasis-free survival.
Our current objectives include :
- Study the molecular organization of the VANGL2/GPCR complex.
- Develop antibodies to detect active GPCR in biopsies.
- Analyze signaling downstream of VANGL2 and GPCR.
- Create pharmacological modulators targeting GPCR.
1.2 MINK1 is a kinase involved in tumor growth and cancer cell dissemination. Our current work includes :
- Development of MINK1 inhibitors.
- Creation of biomarkers to predict inhibitor efficacy.
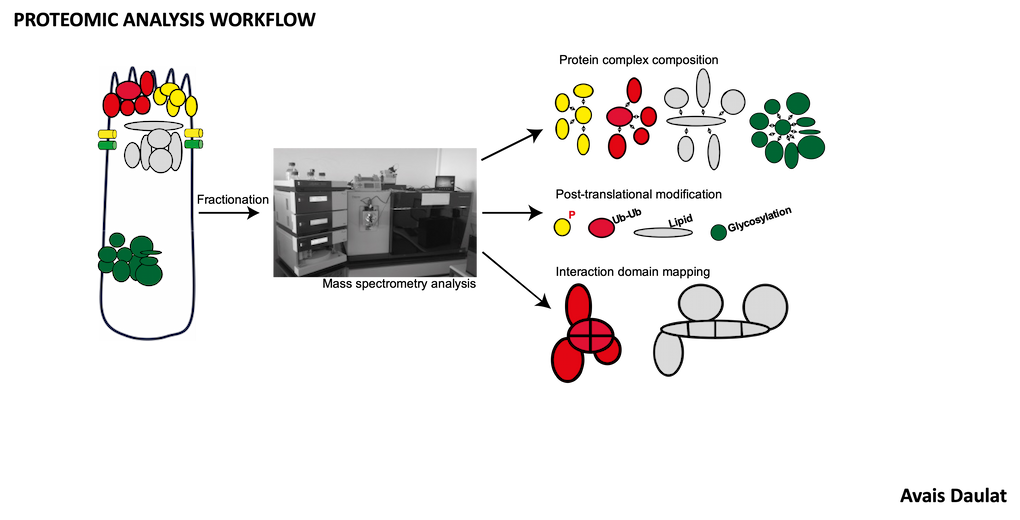
- Analysis of MINK1 signaling by mass spectrometry.
- Evaluation of inhibitors in murine and PDX models of TNBC.
1.3 PTK7 is a pseudo-kinase receptor involved in the WNT/PCP and WNT/β-catenin pathways. We have developed small molecule inhibitors and are working on :
- Explore the structure-activity relationships of inhibitors.
- Test their efficacy in colorectal cancer.
1.4 ADAMTSL5 is a secreted glycoprotein involved in various cancers. Our work shows that its inhibition reduces tumor aggressiveness. We are currently :
- Confirm ADAMTSL5 as a biomarker.
- Let's take a look at its protein complex and mechanism of action.
- Let's study its impact on the tumor microenvironment.
- Let's develop agents for diagnosis and targeting.
This axis aims to explore the involvement of new candidate genes/mechanisms in the dialogue between cancer cells and their microenvironment, in particular with immune cells. It is based on the combination of patient datasets and samples with ex vivo (organoid/tumoroid), single-cell and spatial, and in vivo cancer models.
2.1. Study of immune interactions in TNBC
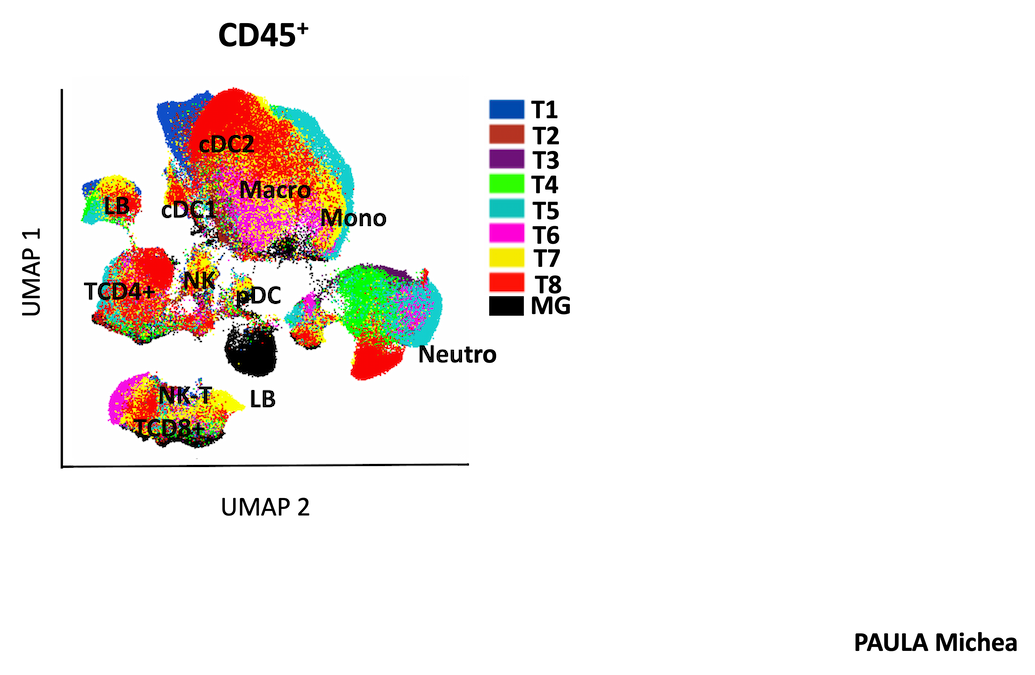
Focusing on TNBC, we use unique in vivo cancer models to explore the composition of the different immune cell types present in the tumor microenvironment and their interaction with cancer cells. We reconstruct and target these interactions in vivo by developing orthotopic models in syngeneic mice with intact immune systems. The results are systematically compared with the contexts observed in patients. In addition, we are exploring longitudinally how distinct anticancer treatments can lead to specific immune remodeling, depending on the type of treatment. These studies are carried out with dynamic rather than static analyses, in order to understand how immune remodeling may influence the most relevant choice of immunotherapies. We anticipate that the acquisition of this knowledge may help in the design of new combinations of anticancer agents and immunotherapies.
2.2. Analysis of early tumor lesions
We have assembled a series of preliminary data indicating that early tumor lesions are characterized by cellular and molecular features that diversify their behavior over time (quiescent versus evolutionary). Focusing on liver cancer, we extend these studies using unique mouse models of spontaneous tumorigenesis, recapitulating the molecular as well as temporal heterogeneity of tumor onset. We have documented the possibility of using photon-counting computed tomography (PC-CT; established by our physics collaborators at CPPM) to longitudinally follow endogenous tumors. Quiescent and evolving lesions are analyzed by spectral cytometry and single cell RNAseq, to uncover molecular and cellular features that diversify an evolving or quiescent behavior of pre-neoplastic lesions. We assess the robustness of the results functionally using ex vivo and in vivo models, and clinically by analyzing patient data and samples.
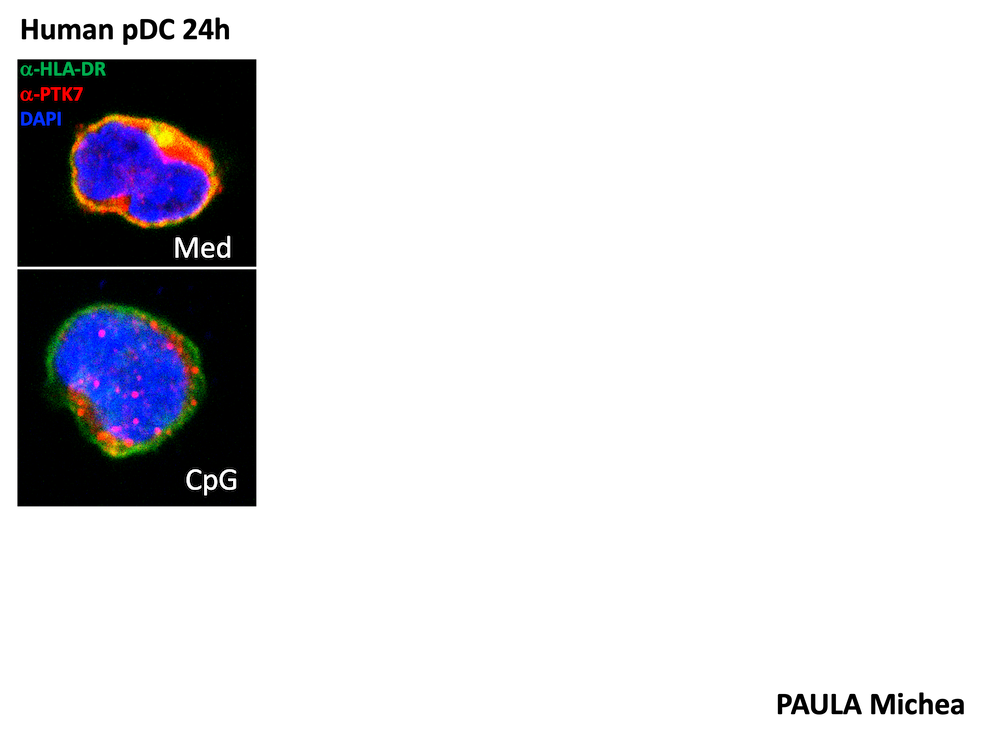
2.3. Role of PTK7 in tumor-host interactions
We recently discovered that PTK7 is expressed in dendritic cells (DCs), particularly Langerhans cells (skin, cutaneous lymph nodes) in mice. In addition, PTK7 expression was also observed in tumor-infiltrating monocytes, pDCs, cDC2 and cDC1 in murine models of melanoma and breast cancer, suggesting that PTK7 may be modulated by the tissue microenvironment.
Using a single mouse model and multiparametric -omics data, we :
- We are studying the physiological function of PTK7 in dendritic cells (DC);
- determine the role of PTK7-expressing DCs in breast cancer. We identified PTK7 expression in colon fibroblasts and increased levels in murine and human CRC. We generated PTK7-deficient mice in a subset of colonic fibroblasts and revealed susceptibility to acute colitis and probable tumorigenesis.
These results indicate an essential role for PTK7 in the dialogue between colonic epithelium and fibroblasts. We are currently :
- document the involvement of PTK7 in the dialogue between colonic epithelium and fibroblasts during tissue homeostasis;
- Molecular and functional characterization of the different cellular compartments regulated by PTK7 using relevant mouse models and human organoids;
- determine the binding partners of PTK7 in the epithelial and fibroblastic compartments using a PTK7-BirA* "knock-in" mouse model in colon carcinogenesis;
- deciphering the contribution of PTK7-expressing fibroblasts to colon cancer tumorigenesis;
- validate PTK7 expression and alterations identified in mice as prognostic markers in colon cancer patients.
Team news
Featured publications
06/2024
Dessaux C, Ganier L, Guiraud L, Borg JP.
04/2024
Walton A, Thomé V, Revinski D, Marchetto S, Puvirajesinghe TM, Audebert S, Camoin L, Bailly E, Kodjabachian L, Borg JP.
11/2022
Sequera C, Grattarola M, Holczbauer A, Dono R, Pizzimenti S, Barrera G, Wangensteen KJ, Maina F.
09/2022
Daulat AM, Wagner MS, Audebert S, Kowalczewska M, Ariey-Bonnet J, Finetti P, Bertucci F, Camoin L, Borg JP.
09/2021
Castellanet O, Ahmad F, Vinik Y, Mills GB, Habermann B, Borg JP, Lev S, Lamballe F, Maina F.
04/2021
Arechederra M, Bazai SK, Abdouni A, Sequera C, Mead TJ, Richelme S, Daian F, Audebert S, Dono R, Lozano A, Gregoire D, Hibner U, Allende DS, Apte SS, Maina F.


Labels, Funding and Partners
Like others, they were part of the team. Thank you to all those who have contributed to CRCM's excellence and impact.