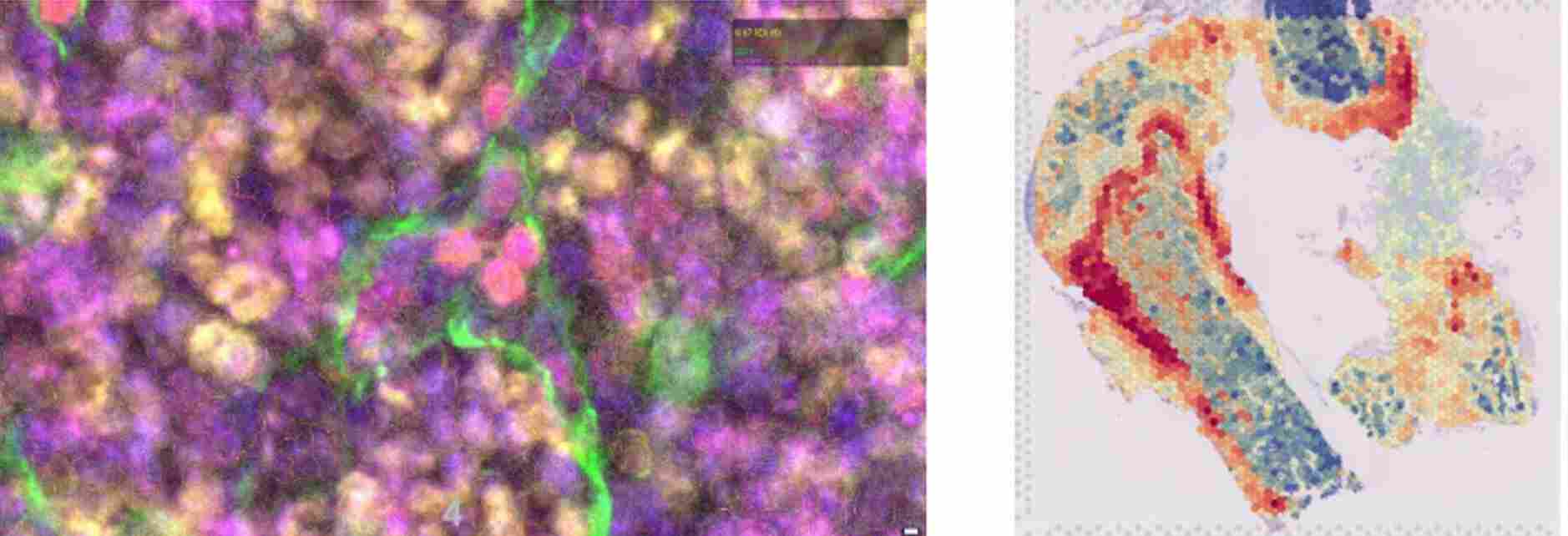
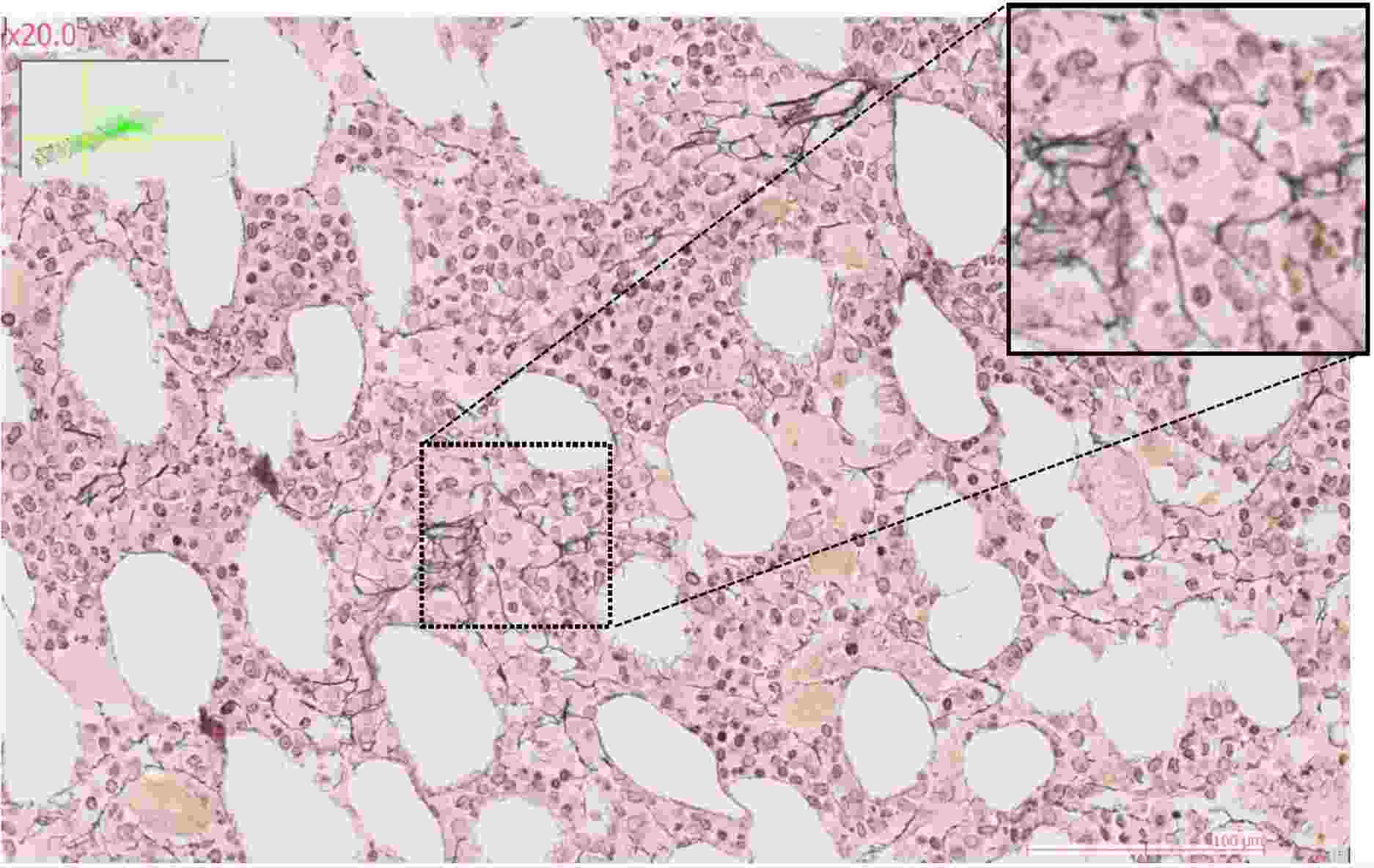
The team's research focuses on characterizing the molecular mechanisms involved in interactions between hematopoietic and stromal cells within bone marrow niches.
Hematopoiesis is the biological mechanism by which hematopoietic stem cells (HSC) give rise to all mature cells in the blood. In adult mammals, hematopoiesis takes place in the bone marrow, and requires interactions between hematopoietic and stromal cells within micro-anatomical units called "niches". Over the past two decades, the concept of "niches" has been extended to the various stages of hematopoietic cell differentiation, suggesting that specific "niches" exist. It is also acknowledged that bone marrow stromal cells play an essential role in the aging of the hematopoietic system and the emergence of myeloid hemopathies, the incidence of which increases with age.
The team's research focuses on characterizing the molecular mechanisms involved in interactions between hematopoietic and stromal cells within bone marrow niches. Our two main lines of research are :
- Identification of biomarkerspredictive of the emergence of myeloid hemopathies and their progression to acute myeloid leukemia (AML).
- Modeling interactionsbetween hematopoietic cells and cells of the bone marrow microenvironment.
The projects relies on the expertise of the team's researchers in immunohematology, cell adhesion biology and the clinical biology of myelodysplastic syndromes (MDS) and myeloproliferative neoplasms (MPN), which can develop into AML in 10% to 30% of cases.
Projects and expertise
The study of biomarkers predictive of the emergence and evolution of myeloid hemopathies in AML is based on histological, phenotypic and molecular analyses of samples from patients with MDS, MDS or AML. We are primarily interested in characterizing the cells at the origin of the various pathologies, which we know accumulate a certain number of gene alterations before progression to overt disease.
These alterations are reflected by changes in gene expression programs in stem cells at the apex of the pathology, resulting in a loss of control of hematopoietic homeostasis. This loss of control involves changes in the spatial organization of the bone marrow in patients with MPN or MDS, some of which progress to AML.
Characterization of diseased stem cells and their microenvironment is at the heart of this line of research, based on data generated within the laboratory and validated (as far as possible) on public data.
The use of label-free imaging technologies, such as quantitative phase imaging or anisotropy measurement (Phasics), enables us to generate quantitative data of a new kind to assess the trabecular bone and collagen fiber content of the bone marrow microenvironment. The ultimate aim of this line of research is to characterize and visualize in situ the interaction networks common to cells of origin of SMP and SMD that evolve into AML compared with cells of origin that do not.
To study stromal changes in pathological conditions, the team uses mouse models of acute myeloid leukemia, focusing on the initial stages of leukemogenesis (bone marrow invasion of less than 20%). These mouse models enable leukemic progression to be monitored using various techniques such as spectral flow cytometry, or imaging techniques such as RNA-scope or confocal microscopy.
In order to understand the changes that occur within the stroma early in leukemic progression, we rely on spatial transcriptomics approaches accompanied by spectral flow cytometry (panels >25 parameters) or single-cell RNA sequencing analyses(scRNAseq).
To this end, the team has developed expertise in spatial transcriptomics at bulk (Visium) or single-cell level (Vizgen and soon Xenium), as well as the requisite bioinformatics skills. The aim of this axis is to identify changes in the bone marrow microenvironment and determine which modifications can be considered as biomarkers or as mechanisms of resistance to treatment. At the same time, we're working to model the bone marrow microenvironment through the fabrication of stromal, endothelial and hematopoietic cell-based bone structures that can be used in vitro or in vivo to improve existing PDX models, which are not very effective for grafting myeloid hemopathies of low aggressiveness or proliferation (MDS, MPN).
Team news
featured publications
09/2024
Grenier JMP, Testut C, Bal M, Bardin F, De Grandis M, Gelsi-Boyer V, Vernerey J, Delahaye M, Granjeaud S, Zemmour C, Spinella JF, Chavakis T, Mancini SJC, Boher JM, Hébert J, Sauvageau G, Vey N, Schwaller J, Hospital MA, Fauriat C, Aurrand-Lions M.
12/2021
Bou-Tayeh B, Laletin V, Salem N, Just-Landi S, Fares J, Leblanc R, Balzano M, Kerdiles YM, Bidaut G, Hérault O, Olive D, Aurrand-Lions M, Walzer T, Nunès JA, Fauriat C.
03/2019
Balzano M, De Grandis M, Vu Manh TP, Chasson L, Bardin F, Farina A, Sergé A, Bidaut G, Charbord P, Hérault L, Bailly AL, Cartier-Michaud A, Boned A, Dalod M, Duprez E, Genever P, Coles M, Bajenoff M, Xerri L, Aurrand-Lions M, Schiff C, Mancini SJC.
09/2018
Aknoun, S., Aurrand-Lions, M., Wattellier, B., and Monneret, S.
12/2017
De Grandis M, Bardin F, Fauriat C, Zemmour C, El-Kaoutari A, Sergé A, Granjeaud S, Pouyet L, Montersino C, Chretien AS, Mozziconacci MJ, Castellano R, Bidaut G, Boher JM, Collette Y, Mancini SJC, Vey N, Aurrand-Lions M.
06/2015
Cartier-Michaud A, Bailly AL, Betzi S, Shi X, Lissitzky JC, Zarubica A, Sergé A, Roche P, Lugari A, Hamon V, Bardin F, Derviaux C, Lembo F, Audebert S, Marchetto S, Durand B, Borg JP, Shi N, Morelli X, Aurrand-Lions M.


Labels, Funding and Partners
Like others, they were part of the team. Thank you to all those who have contributed to CRCM's excellence and impact.